Multidisciplinary Research Project: Fraction Library
ECR Project Leaders:
Iva Veseli and Yuting Zhu
PI Project Leader:
Art Edison
Research Project Description
We are creating a knowledge base of microbially-produced compounds as a reference library for the microbe-metabolite network of the surface ocean.
The C-CoMP Fraction Library leverages the analytical strengths of liquid chromatography-mass spectrometry (LC-MS and LC-MS/MS) and nuclear magnetic resonance (NMR) spectroscopy, enabled by a novel derivatization method that captures small polar compounds from marine media or seawater by attaching a benzoyl group to alcohols and amines (Widner et al. 2021, Ferrer-González et al. 2021). This approach leverages the strengths of both analytical methods to annotate unknown metabolites with a high level of confidence. The fraction library will be available in formats that can be readily used, shared, and updated by the scientific community.
Building the C-CoMP Fraction Library
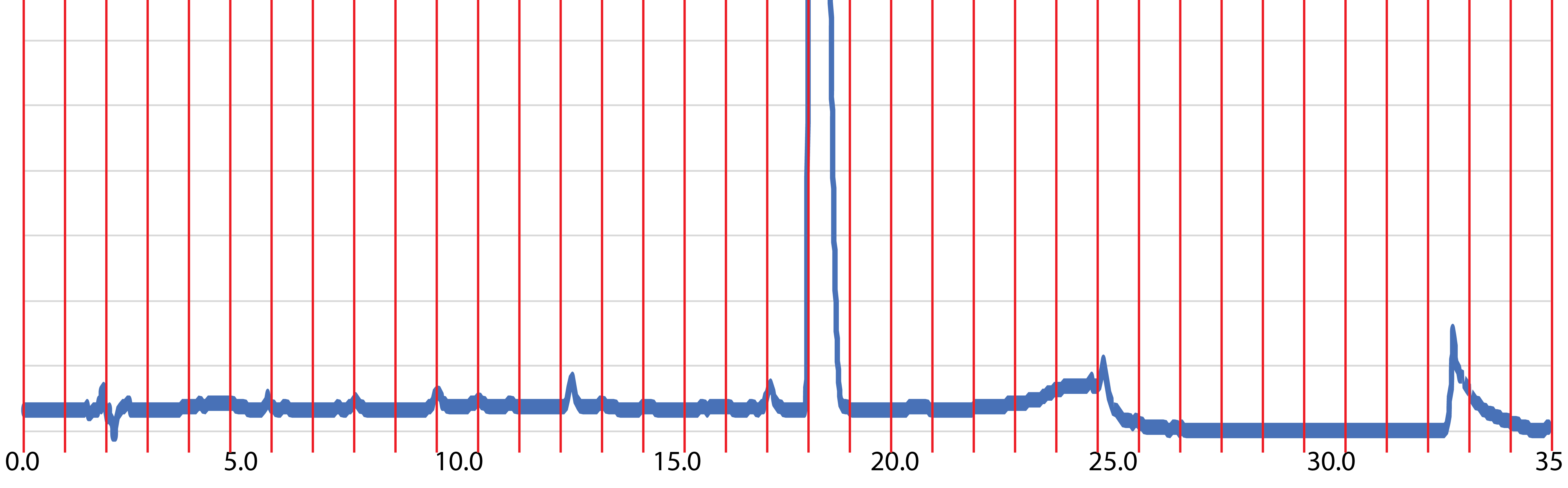
Fraction library construction is beginning with exometabolites from axenic cultures of marine diatom Thalassiosira pseudonana CCMP1335. An enriched pool of small polar compounds is obtained through benzoyl chloride derivatization of spent medium followed by concentration via solid phase extraction resin (Widner et al. 2021). The compound mixture is divided into fractions by high performance liquid chromatography (HPLC) (Fig. 1), with aliquots of each fraction analyzed by both LC-MS and NMR.
Gaining Biological Insight from Metabolite Fraction Libraries
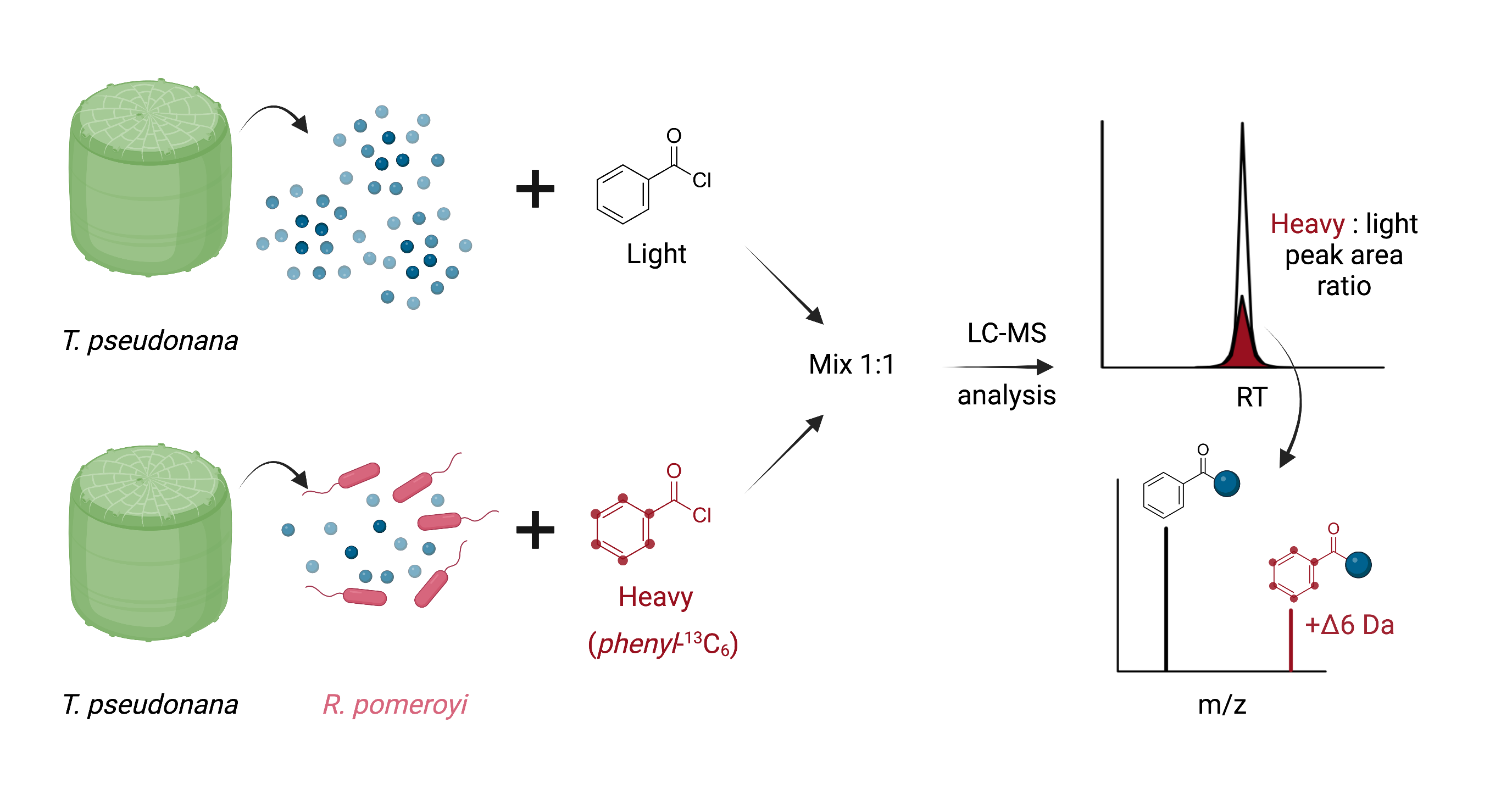
The fraction libraries are built from derivatized exometabolites, and require new types of biological/physiological experiments in order to utilize the rich data. The first experiment will be to compare an axenic exometabolite culture from T. pseudonana CCMP 1335 with and without addition of a heterotrophic bacterium. After the incubation, the culture without bacteria will be derivatized with unlabeled benzoyl chloride and the one with bacteria will be derivatized with isotopically-enriched benzoyl chloride. Next, the two cultures are mixed and extracted together, and the material analyzed by LC-MS. The chromatographic retention times will be similar between the isotopically-enriched and unlabeled derivatives, but the m/z values will differ. Metabolites that significantly change intensities in this experiment are then compared with the fraction library from T. pseudonana that was derivatized with the same protocol, enabling a link between one- and two-dimensional NMR data with LC-MS and LC-MS/MS data for more confident metabolite identification.
References
Widner, B., M. C.Kido Soule, F. X.Ferrer-González, M. A. Moran, and E. B. Kujawinski. 2021. Quantification of amine-and alcohol-containing metabolites in saline samples using pre-extraction benzoyl chloride derivatization and ultrahigh performance liquid chromatography tandem mass spectrometry (UHPLC MS/MS). Analytical Chemistry 93: 4809-4817.
Ferrer-González, F. X., B. Widner, N. R. Holderman, J, Glushka, A. Edison, E. B. Kujawinski, and M. A. Moran. 2021. Resource partitioning of phytoplankton metabolites that support bacterial heterotrophy. The ISME Journal 15: 762-773.
