Multidisciplinary Research Project: Phytoplankton Exometabolites and Proteins
ECR Project Leaders:
Yuting Zhu and Hanna Anderson
PI Project Leader:
Sonya Dyhrman
Research Project Description
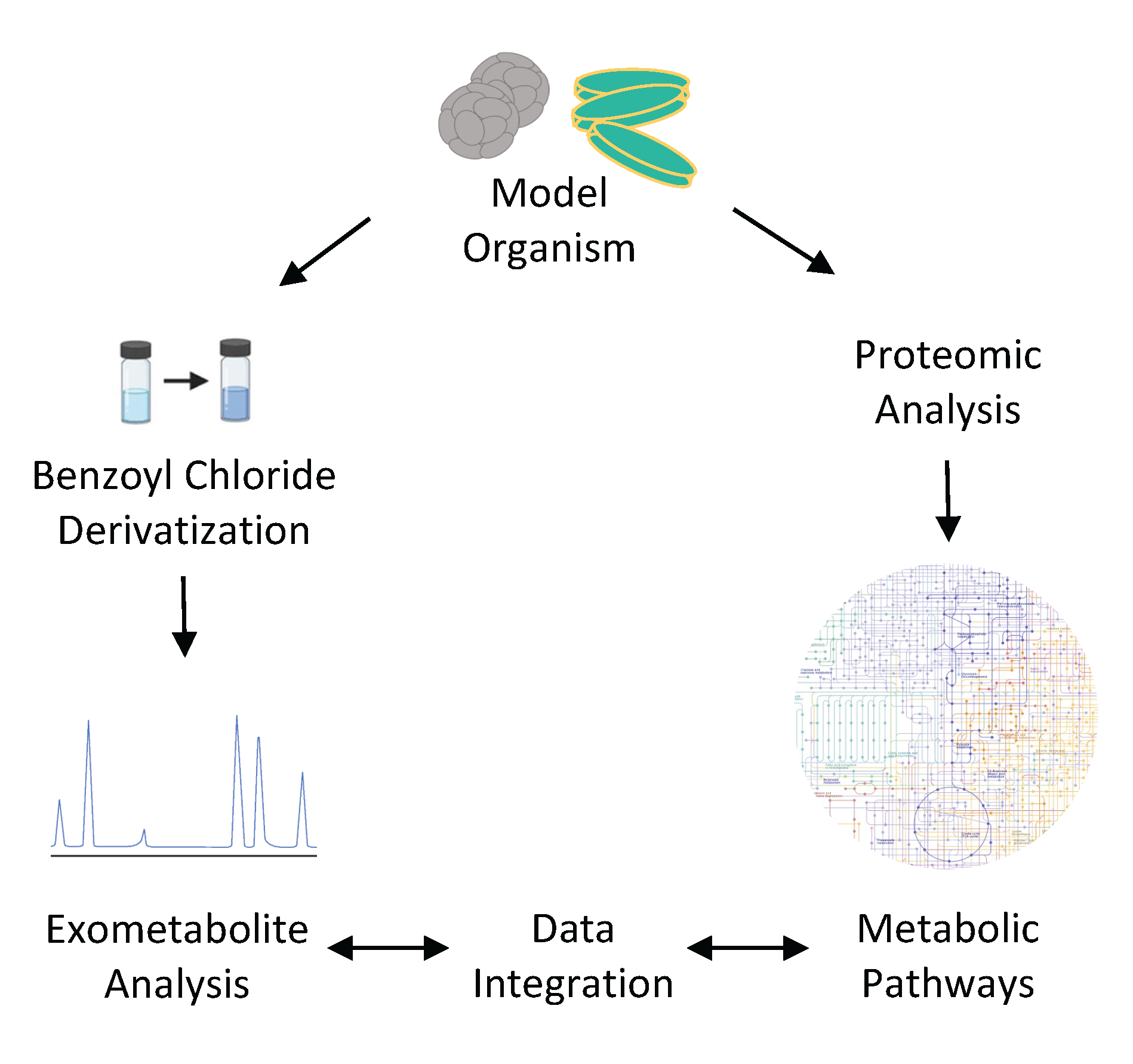
What are the chemical currencies of our microbial oceans? We are addressing this core C-CoMP question by examining the exometabolites released from representative taxa of marine phytoplankton. These collections of organic compounds released from marine phytoplankton are poorly characterized, largely due to the difficulties in measuring polar metabolites in saline water. In this work, we are identifying extracellular metabolites and linking them with their production pathways using parallel proteomic analysis. Knowledge of taxon-specific biosynthesis pathways will help us link exometabolites observed in field studies to their producers.
Phytoplankton Screening
Samples were obtained from axenic cultures of 6 phytoplankton species representing key aspects of functional and taxonomic diversity (Diatom: Thalassiosira pseudonana CCMP1335, Picocyanobacteria: Synechococcus WH8102, Prochlorococcus marinus MIT9301, Diazotroph: Crocosphaera watsonii WH8501, Coccolithophore: Emiliania huxleyi CCMP371, and picoeukaryote: Micromonas commoda RCC299). Exometabolites were characterized with LC-MS analysis after derivatization with benzoyl chloride (Widner et al., 2021). Proteomes were analyzed with LC-MS using data-independent acquisition (DIA) for high-precision analysis of all peptides.
Data Integration
To facilitate comparisons across taxa, data sharing, and C-CoMP’s data integration efforts, these data will be represented in Digital Microbes, gene-centric datasets for individual marine microbes that integrate genome, proteome, and metabolite profiles. We will (1) store analyzed proteome data per gene in data tables in anvi’o, and (2) export reaction networks from the genomes for comparison with exometabolites measured using LC-MS.
References
Widner, B., Kido Soule, M.C., Ferrer-González, F.X., Moran, M.A. & Kujawinski, E.B. 2021. Quantification of amine-and alcohol-containing metabolites in saline samples using pre-extraction benzoyl chloride derivatization and ultrahigh performance liquid chromatography tandem mass spectrometry (UHPLC MS/MS). Analytical Chemistry 93:4809-4817.
