Multidisciplinary Research Project: Digital Microbes
The results of this project are now published! Check out the following publication:
Veseli, I., DeMers, M. A., Cooper, Z. S., Schechter, M. S., Miller, S., Weber, L., Smith, C. B., Rodriguez, L. T., Schroer, W. F., McIlvin, M. R., Lopez, P. Z., Saito, M., Dyhrman, S., Eren, M. A., Moran, M. A., & Braakman, R. (2024). Digital Microbe: a genome-informed data integration framework for team science on emerging model organisms. Scientific Data, 11(1), 967. doi: https://doi.org/10.1038/s41597-024-03778-z.
ECR Project Leaders:
Zac Cooper and Matt Schechter
PI Project Leader:
Meren
Research Project Description
We are building resources that allow effective sharing of common digital datasets for marine microbial taxa. A ‘Digital Microbe’ organizes and integrates 'omics and ‘omics-related datasets specific to a microbial taxon in an easily-accessible, interoperable, and reproducible manner. As a National Science Foundation Science and Technology Center (STC) committed to open science, we follow FAIR data principles.
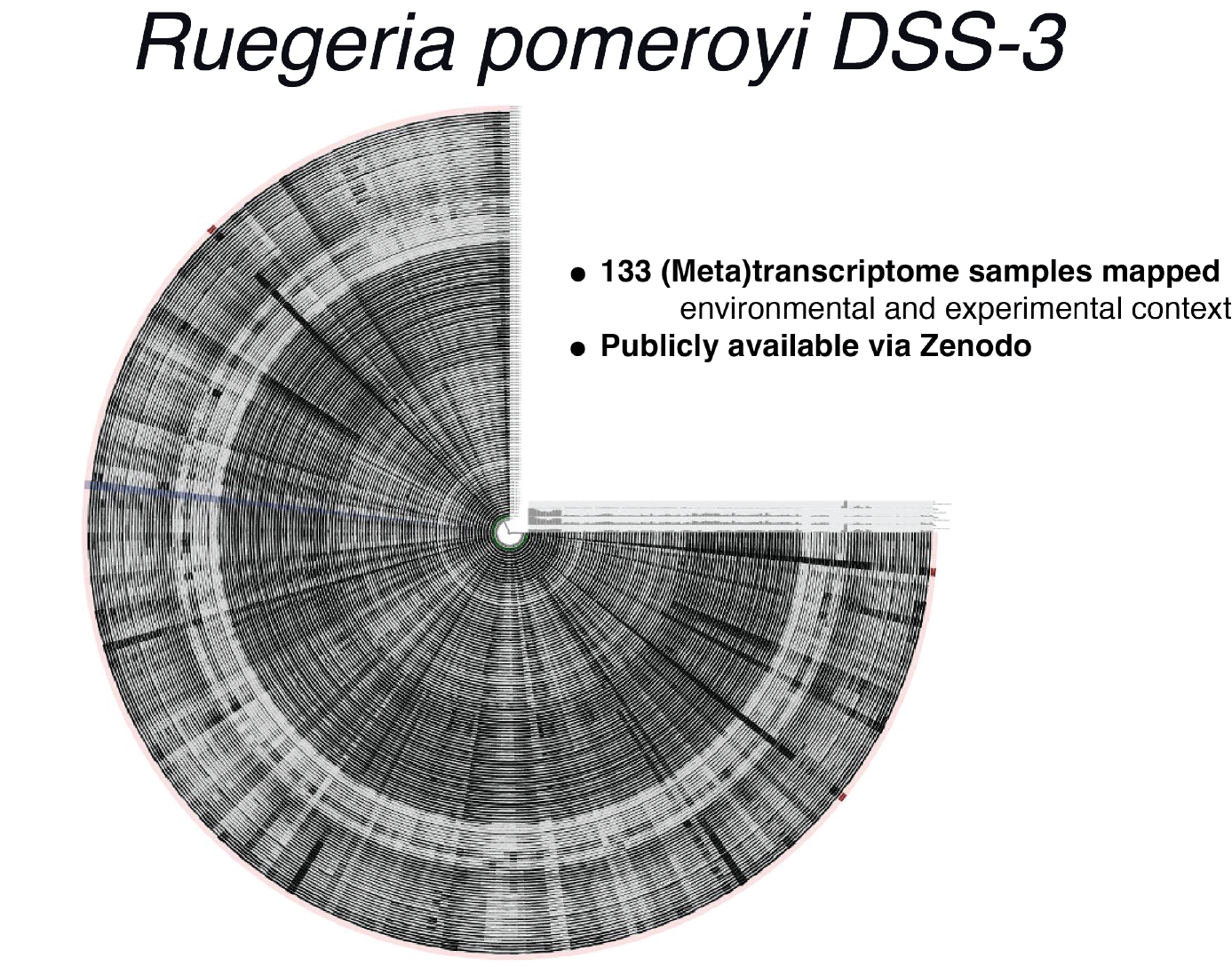
The Ruegeria pomeroyi DSS-3 Digital Microbe
One of the two model marine bacteria central to C-CoMP research is Ruegeria pomeroyi DSS-3, an Alphaproteobacterium in the Rhodobacterales family and a member of the Roseobacter group. This copiotrophic marine bacterium was isolated from coastal seawater in the 1990s and its genome was sequenced in 2004 (Moran et al, 2004). Since then, gene calls have been continually curated to incorporate new knowledge reflecting the ecology and physiology of this microbe. In this project, the well-curated genome is the coordination framework for a collection of data types that together comprise the Digital Microbe.
To build the R. pomeroyi Digital Microbe we used the community-driven, open-source software platform anvi’o. The first version of this reproducible R. pomeroyi data resource is available on Zenodo, and includes a contigs-db and a profile-db, which are stand-alone SQLite databases compatible with the anvi'o software ecosystem for a wide range of analyses. The R. pomeroyi Digital Microbe gives access to the genome’s curated open-reading frames and functional annotations. In addition, it makes available the results of specific experimental observations, such as transcriptomes, and provides information on single-gene mutants. The technical details of this format and the reproducible workflow that generated the R. pomeroyi files are available in a C-CoMP blog post by Iva Veseli. Our first use of this resource has focused on the classification of R. pomeroyi gene expression patterns under various conditions, and will serve to bring together groups using the R. pomeroyi model inside and outside of C-CoMP.
The Alteromonas Digital Microbe
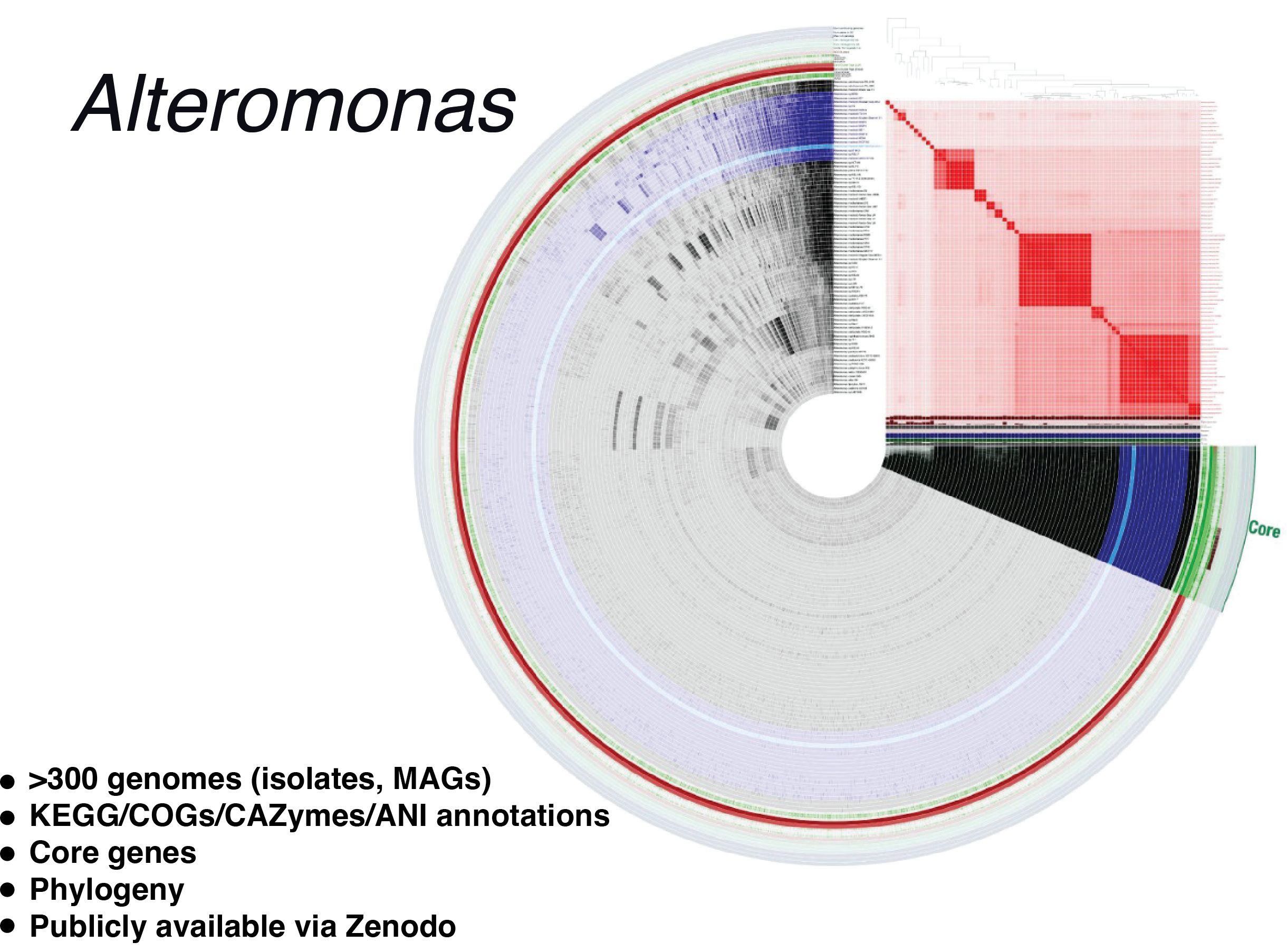
C-CoMP’s second focal marine bacterium is Alteromonas macleodii MIT1002, a ‘helper’ bacterium that participates in a mutualistic relationship with the ubiquitous ocean cyanobacterium Prochlorococcus (Biller et al. 2015). The Alteromonas Digital Microbe is designed for studies of the species’ pangenome (i.e., the full complement of genes carried by members of a species) rather than a single genome. This pangenomic Alteromonas Digital Microbe is accessible and queryable using the anvi’o platform, and integrated ‘omics datasets streamline downstream analyses and visualization. Our first use of this database is analysis of pangenome phylogeny as a function gene repertoire, and characterization of abundance and strain-level divergence in ocean communities by metagenomic mapping. The Alteromonas Digital Microbe is available on Zenodo.
A General Framework for Microbial Biology Teams
The anvi’o-based Digital Microbe datasets offer an open resource for sharing curated and versioned genomes and associated functional data, ensuring the availability of a common bioinformatic foundation for a scientific team or community. Digital Microbes are built with standard features of the anvi’o open source bioinformatic platform, stored as SQLite databases, and can be readily shared and updated by community members. A eukaryotic Digital Microbe platform is planned for the near future.
Learn more:
You can learn more about Digital Microbes by reading the preprint entitled "Digital Microbe: a genome-informed data integration framework for collaborative research on emerging model organisms" and the preLights article..
References
Moran, M.A., Buchan, A., González, J.M., Heidelberg, J.F., Whitman, W.B., Kiene, R.P., Henriksen, J.R., King, G.M., Belas, R., Fuqua, C. and Brinkac, L. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910-913.
Eren, A.M., Esen, Ö.C., Quince, C., Vineis, J.H., Morrison, H.G., Sogin, M.L. and Delmont, T.O. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319.
Biller, S. J., A. Coe, A.-B. Martin-Cuadrado, and S. W. Chisholm. 2015. Draft genome sequence of Alteromonas macleodii Strain MIT1002, Isolated from an Enrichment Culture of the Marine Cyanobacterium Prochlorococcus. Genome Announcements 3:e00967-15.
