Data Access
Data generated by C-CoMP laboratories is stored in repositories that provide DOI numbers to ensure persistent access to our products, unless otherwise indicated. All repositories (see below) provide immediate access to the broader community. Metabolite data will be deposited in either MetaboLights or the Metabolomics Workbench. Field data from the Bermuda Atlantic Time-series (BATS) is stored at the BATS ftp website and at the NSF-supported Biological & Chemical Oceanography Data Management Office (BCO-DMO) repository. Processed protein data is stored at BCO-DMO and accessed through the Ocean Protein Portal. Proteomics raw mass spectral data is submitted to ProteomeXchange via Massive or Pride. Sequence data is stored at NCBI on the Sequence Read Archive (SRA), iMicrobe, and/or iVirus. We use Zenodo to share versioned, intermediate data products. Raw model data will be stored on local servers initially. For CESM results, we will adhere to their data policy, which includes data release within one year of generation. In general, C-CoMP will meet the NSF guidelines on data release within 2 years of generation but will release any hardened data products before then, if possible. All software development efforts in C-CoMP will follow open source software development practices, will be licensed through General Public License, and the source code of stable releases as well as the development branches will be accessible to the community through GitHub repositories. Stable releases of our software will also be available as Conda packages through the Anaconda Package Repository and as Docker containers through the Docker Hub for platform independent, easy-to-install or easy-to-run scenarios.
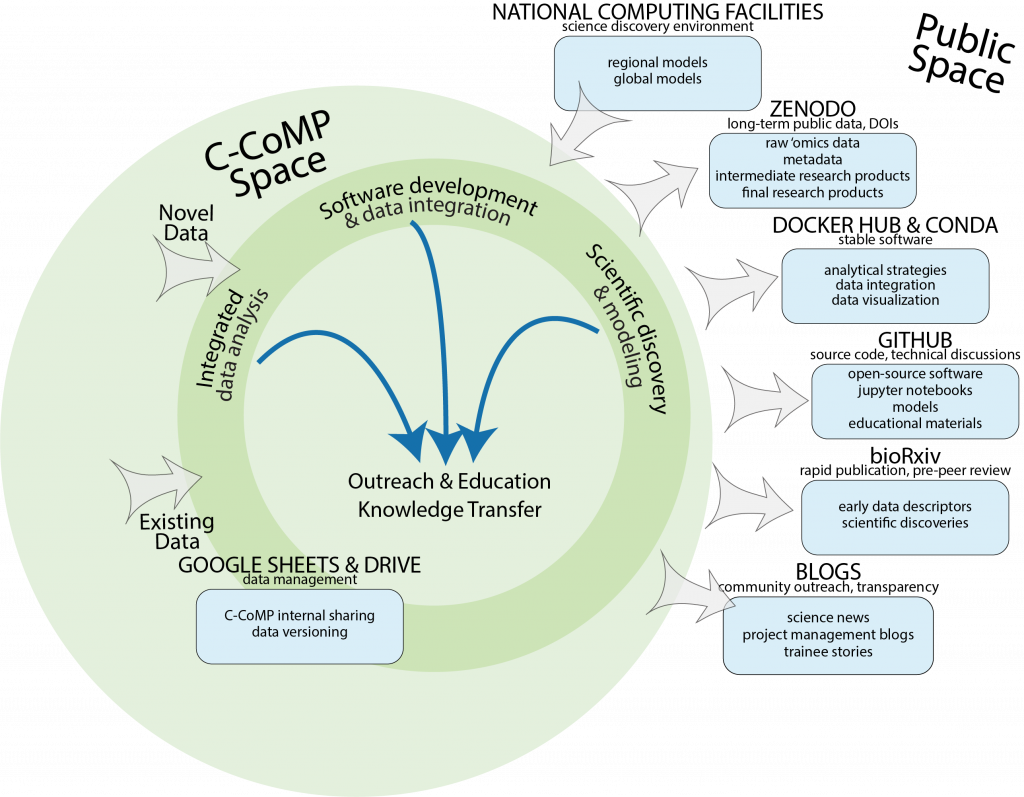
The figure below displays strategies for C-CoMP knowledge transfer within the ‘C-CoMP space’ and with the ‘Public space’. C-CoMP contributes to education outreach and knowledge transfer through research that includes integrated data analysis, software development and integration, and scientific discovery and modeling. Novel and existing data are inputs that enable this research. Data management, including internal sharing of datasets and versioning within the shared C-CoMP Google Drive, facilities the efficiency and speed of data sharing. C-CoMP knowledge transfer to the ‘public space’ includes:
- long-term public data (raw ‘omics data, metadata, intermediate research products, and final research products) with assigned digital object identifiers (DOI’s)
- stable software designed for analyzing, integrating, and visualizing data shared through the Docker Hub or Conda
- source code and technical discussions for open-source software, models, and educational materials shared through GitHub
- rapid publication and pre-peer review of manuscripts for providing early data descriptors and scientific discoveries via BioRxiv or other pre-print journals.
- blog posts sharing science news, project management ideas, and trainee stories for community outreach and transparency.
| Data type | Repository/website |
|---|---|
| Raw metabolite data | MetaboLights and/or Metabolomics Workbench |
| Field data | BATS, BCO-DMO |
| Processed protein data | BCO-DMO via Ocean Protein Portal |
| Raw proteomics data | ProteomeXchange |
| Genomic Sequence data (16S rRNA, Whole Genome Sequencing, Shotgun Metagenomics) | NCBI SRA, iMicrobe, iVirus |
| Open source software | Github, Anaconda, Docker |
| CESM results | TBD |
