Arrayed Mutant Libraries
Description
A mutant library of bacterium Ruegeria pomeroyi DSS-3, a C-CoMP focal strain, was created by individually arraying random transposon mutants in multi-well plates. Visit the following link to access the catalog of arrayed mutants spreadsheet. An earlier version of this catalog had incorrect gene identifications for about 20% of the mutant locations, and we apologize for any issues it may have caused.
Pooled mutant libraries (i.e., a mixed pool of thousands of transposon mutants) are available for R. pomeroyi and for Alteromonas sp. MIT1002. These are suitable for RB-TnSeq studies (Cain et al., 2020).
The mixed mutant pools are available upon request. Copies of the arrayed library may be available, depending on supplies. Please direct inquiries to Mary Ann Moran (mmoran@uga.edu).
Arrayed Mutant Library Construction
Randomly barcoded transposon mutants were constructed by introducing a Tn5 barcoded library into R. pomeroyi DSS-3 cells which inserted randomly into cell DNA (Wetmore et al. 2015). The genes disrupted by the transposons were linked to their unique barcodes by sequencing. Tens of thousands of unique transposon mutants exist together in a single mixed pool at this stage; these can serve as starting material for RB-TnSeq analysis (Wetmore et al. 2015). To array individual mutants, colonies were isolated from the mixed mutant pool on solid medium and distributed into 384 well microplates. The mutant located in each well was determined by PCR using unique combinations of indexed primers that indicate the row and column positions (Mejia et al., 2022). The completed RB-TnSeq arrayed library contains 4,991 mutants arrayed in 13 microplates that cover 2,977 genes in the R. pomeroyi genome (70% of the 4,288 genes with 1 or 2 mutants per gene).
Using the Mutant Library for Transporter Annotation
To demonstrate use of the arrayed library to improve understanding of metabolite-microbe networks, a set of 156 influx transporter mutants was assembled from the R. pomeroyi arrayed RB-TnSeq library and used in high-throughput screens against possible cognate substrates. Mutants were inoculated into medium containing one potential substrate; in total, 70 metabolites were screened. A mutant exhibiting a growth defect on a metabolite was further screened by chemical analysis to confirm it was no longer able to draw down the compound in a growth medium (Fig. 1). The screens experimentally verified substrates of thirteen R. pomeroyi transporter systems (Schroer et al., 2023).
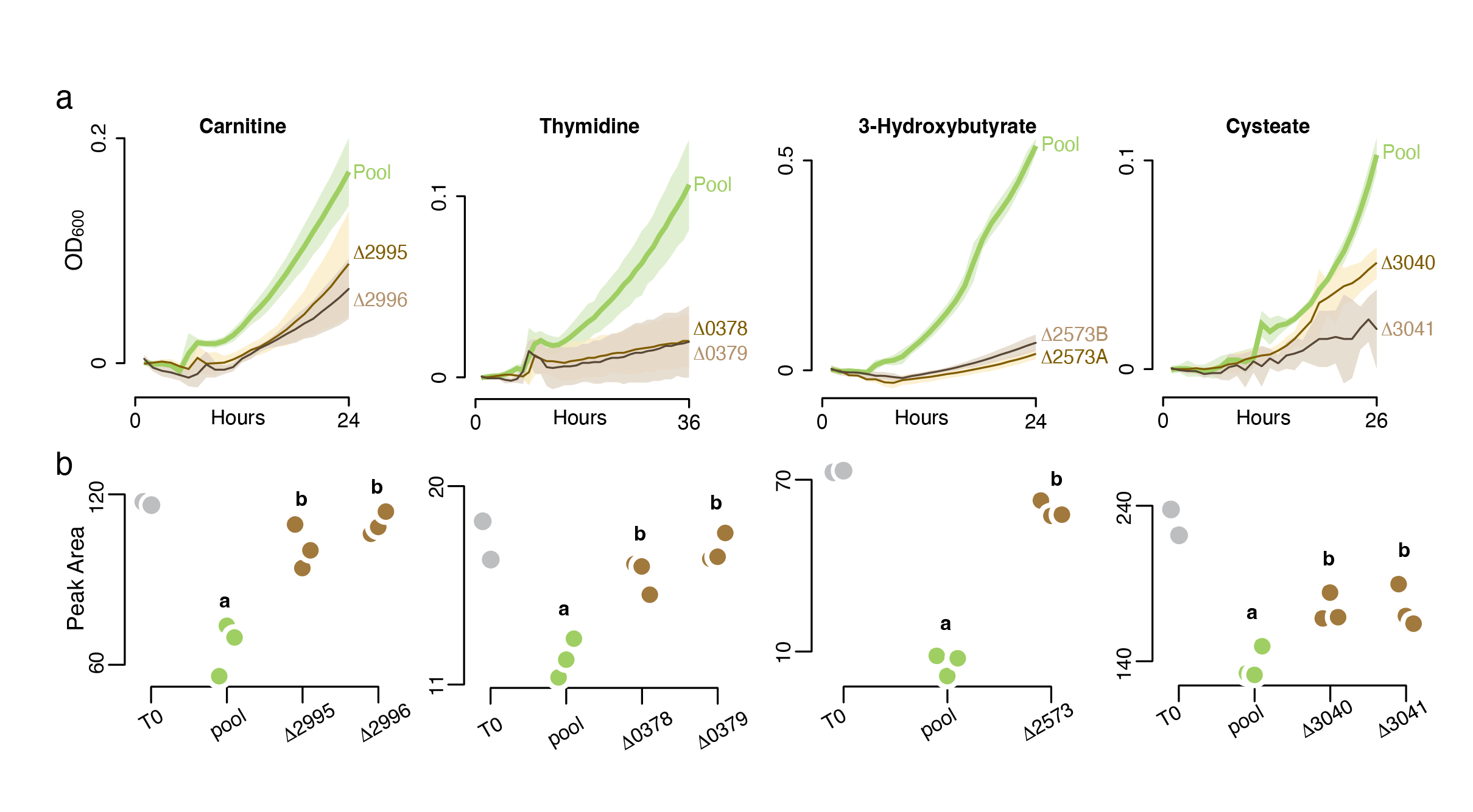
The Metabolite Landscape of a Coastal Phytoplankton Bloom
The new transporter annotations were used to explore the availability of labile dissolved organic compounds during a natural phytoplankton bloom. R. pomeroyi DSS-3 was introduced into a dinoflagellate-dominated bloom in Monterey Bay on 14 dates in the fall of 2016 (Nowinski et al., 2021) and the bacterium’s response to bloom conditions was assessed from gene expression patterns. Reanalysis of the expression dataset given new knowledge of transporter cognate substrates revealed that citrate, 3-hydroxybutyrate, taurine, and DMSP (dimethylsulfoniumpropionate) were among the most available compounds to R. pomeroyi when introduced into the bloom community. DHPS (dihydroxypropanesulfonate) and cysteate were among the least available (Schroer et al., 2023) (Fig. 2). These results are consistent with chemical analyses of phytoplankton endometabolomes which show high concentrations of taurine and DMSP in dinoflagellate cells (Durham et al. 2021; Matrai and Keller 1994) yet low or zero concentrations of DHPS and cysteate (Durham et al. 2021).
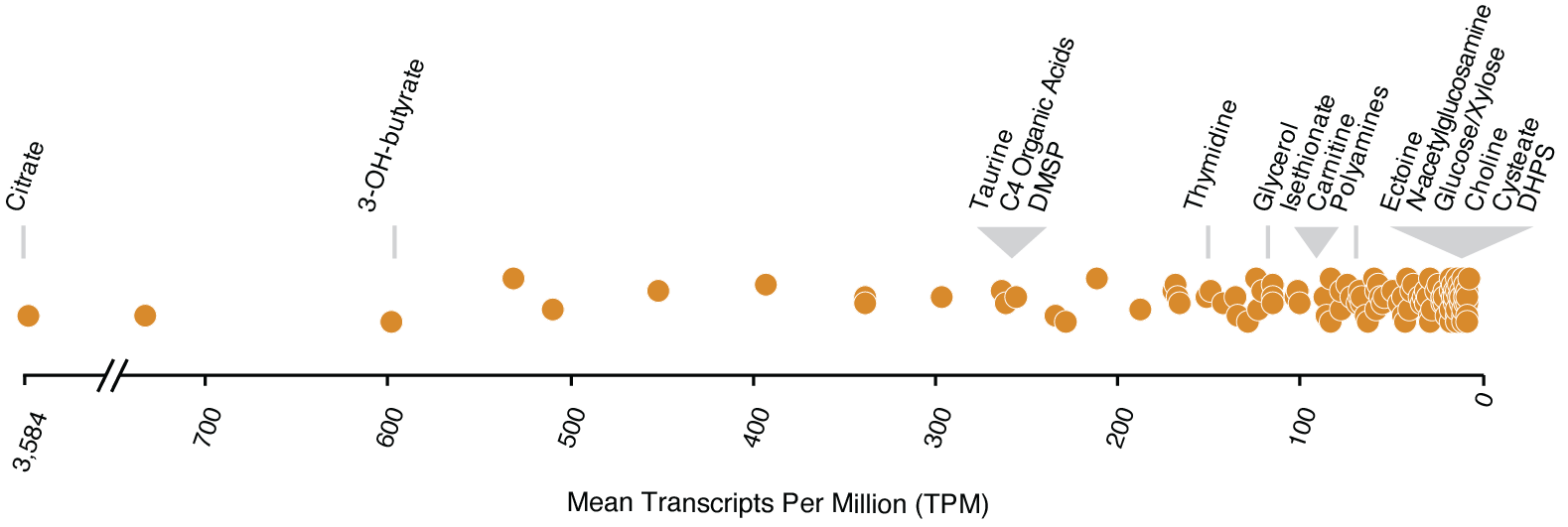
Tools for experimental confirmation of gene function have been limited primarily to cultured bacteria for which genetic systems are available, excluding most of the diversity in Earth’s microbiomes. Construction of arrayed mutant libraries requires only a minimal genetic system sufficient to introduce small DNA fragments (transposons) and an enzyme (transposase) into bacterial cells, making it a low bar for genetic manipulations of environmental strains. Improved functional annotation of the genes mediating marine metabolite production and consumption will build knowledge of carbon flux and fate in microbial ecosystems.
References
Cain, A.K., Barquist, L., Goodman, A.L., Paulsen, I.T., Parkhill, J., and van Opijnen, T. 2020. A decade of advances in transposon-insertion sequencing. Nature Reviews Genetics 21:526-40.
Durham, B.P., Boysen, A.K., Carlson, L.T., Groussman, R.D., Heal, K.R., Cain, K.R., et al. 2019. Sulfonate-based networks between eukaryotic phytoplankton and heterotrophic bacteria in the surface ocean. Nat Microbiol. 4:1706-15.
Matrai, P.A., and Keller, M.D. 1994. Total organic sulfur and dimethylsulfoniopropionate in marine phytoplankton: intracellular variations. Marine Biology 119: 61-68.
Mejia, C., Rodriguez, L.T., Poudel, R., Ellington, A., Rivers, A., and Reisch, C. 2022. An arrayed transposon library of Ruegeria pomeroyi DSS-3. bioRxiv :2022.09.11.507510.
Nowinski, B., and Moran, M.A. 2021. Niche dimensions of a marine bacterium are identified using invasion studies in coastal seawater. Nat Microbiol. 6:524-32.
Schroer, W.F., Kepner, H.E., Uchimiya, M., Mejia, C., Rodriguez, L.T., Reisch, C., and Moran, M.A. Function and importance of marine bacterial transporters of plankton exometabolites. ISME Communications. In press.
Wetmore, K.M., Price, M.N., Waters, R.J., Lamson, J.S., He, J., Hoover, C.A., et al. 2015. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio. 6:e00306-15.
