Network Sensitivity and Feedbacks on Climate
Research Theme Summary:
C-CoMP is working to uncover the principles that control the fate of labile dissolved organic carbon (DOC) on regional to global scales and assess sensitivity to climate feedbacks.
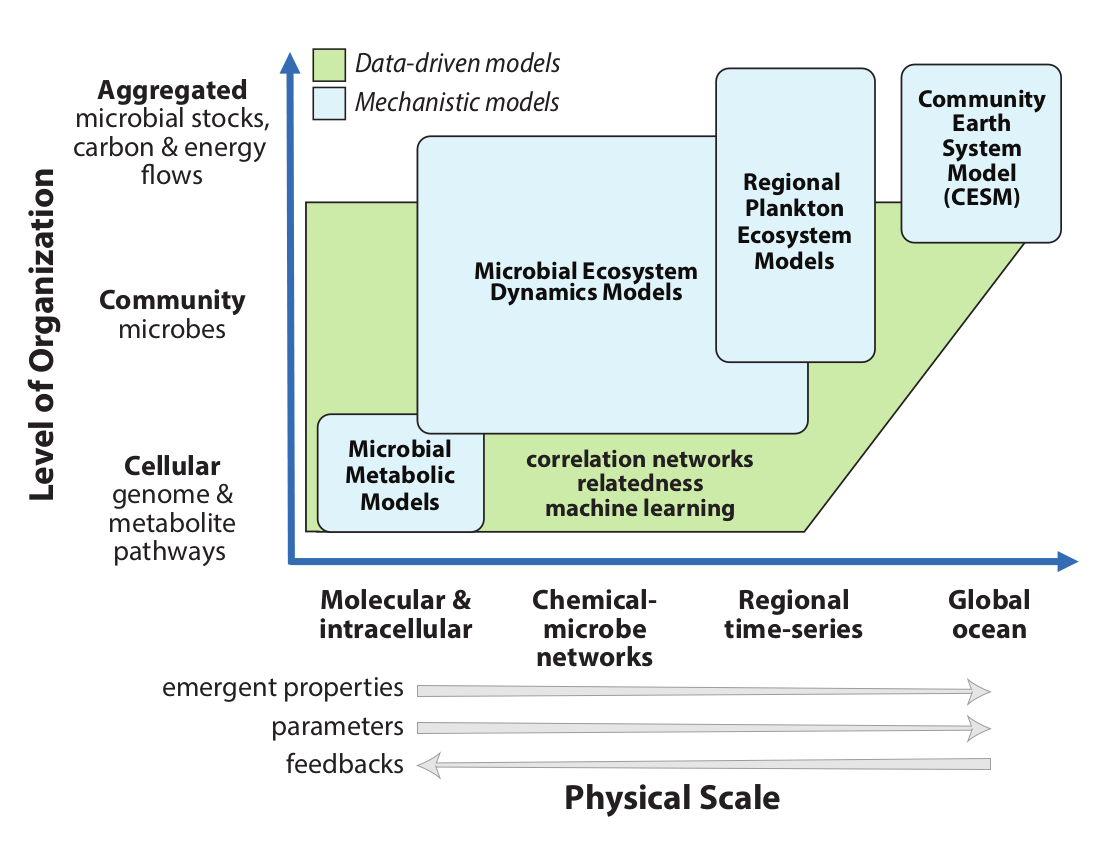
Modeling across scales of complexity
We will formulate a new resource allocation model for marine microbes by exploiting the conceptual convergence between the allocation approaches in commonly used phytoplankton-zooplankton-nutrient-detritus (PZND) models and the aggregated output from flux balance analysis (FBA) models. We will employ a systematic approach to model development and evaluation that is grounded in theory and encapsulates rules of the chemical-microbe network. We will search for predicted emergent feedbacks across levels of biological organization and characterize functional responses of phenotypic metrics - carbon excretion, carbon use efficiency, and carbon transformation rates – to improve parameterizations. Representations of chemical-microbe interactions will focus on responses to warming, stratification, nutrient limitation, and ocean acidification. We will determine the sufficient, but computationally tractable, number of aggregated microbe groups and labile metabolites to include in carbon models; identify slow-evolving community metabolic pathways that must be treated explicitly (prognostically) compared to faster processes and quasi-steady state stocks that can be approximated implicitly from overall system state; and determine functional forms, saturation, and limitation parameters.
Regional biogeochemical models
Translation of C-CoMP science to regional scales requires that details of the chemical-microbe network be simplified in the PZND - biogeochemical modules. C-CoMP efforts will build on the multi-species models from flux balance analysis (FBA) COMETS to explicitly address the composition, characteristics, and rates of metabolic currency exchange between microbes that predict outcomes of the fast and outsized carbon loop linking the surface ocean and atmospheric reservoirs. These model approaches will be evaluated in regional 1-D biogeochemical simulations against field data collected at the ocean time-series sites (Bermuda Atlantic Time-series and Martha's Vineyard Coastal Observatory). Results will in turn be used to predict the distribution of metabolic pathways and functions in the ocean, which can be examined using metagenomics.
Global Biogeochemical Models
The new, more advanced model treatments of chemical-microbe dynamics developed earlier and tested against regional field data will be incorporated into PZND global ocean simulations to explore carbon cycle ramifications. We will use the ocean biogeochemical component of the Community Earth System Model (CESM) for these experiments because CESM is a National Science Foundation (NSF)-supported activity with an open source code, publicly available reference simulations, sophisticated model analysis toolkits, and substantial model development and user communities. Although more biologically sophisticated modeling approaches are used in some large-scale ocean models to incorporate explicit competition and selection among plankton species and for trait-based approaches, the PZND-based CESM is used widely for international model comparisons of historical and future climate change and human carbon cycle perturbations. As with regional models, results from global models will be used to make predictions on the distribution of pathways that can be examined using metagenomics.
Knowledge Transfer
This research advances the following Knowledge Transfer aims:
- Ocean data ingestion - supporting model development and testing
- Molecular biology - investigating model predictions of links between microbial traits and carbon fate
