Multidisciplinary Research Project: Bacterial Carbon Use Efficiency
ECR Project Leaders:
Helen Scott
PI Project Leader:
Daniel Segrè
Research Project Description
We are integrating genome scale modeling and physiological measurements to understand the roles of core metabolism and substrate chemistry in determining bacterial carbon use efficiency (CUE).
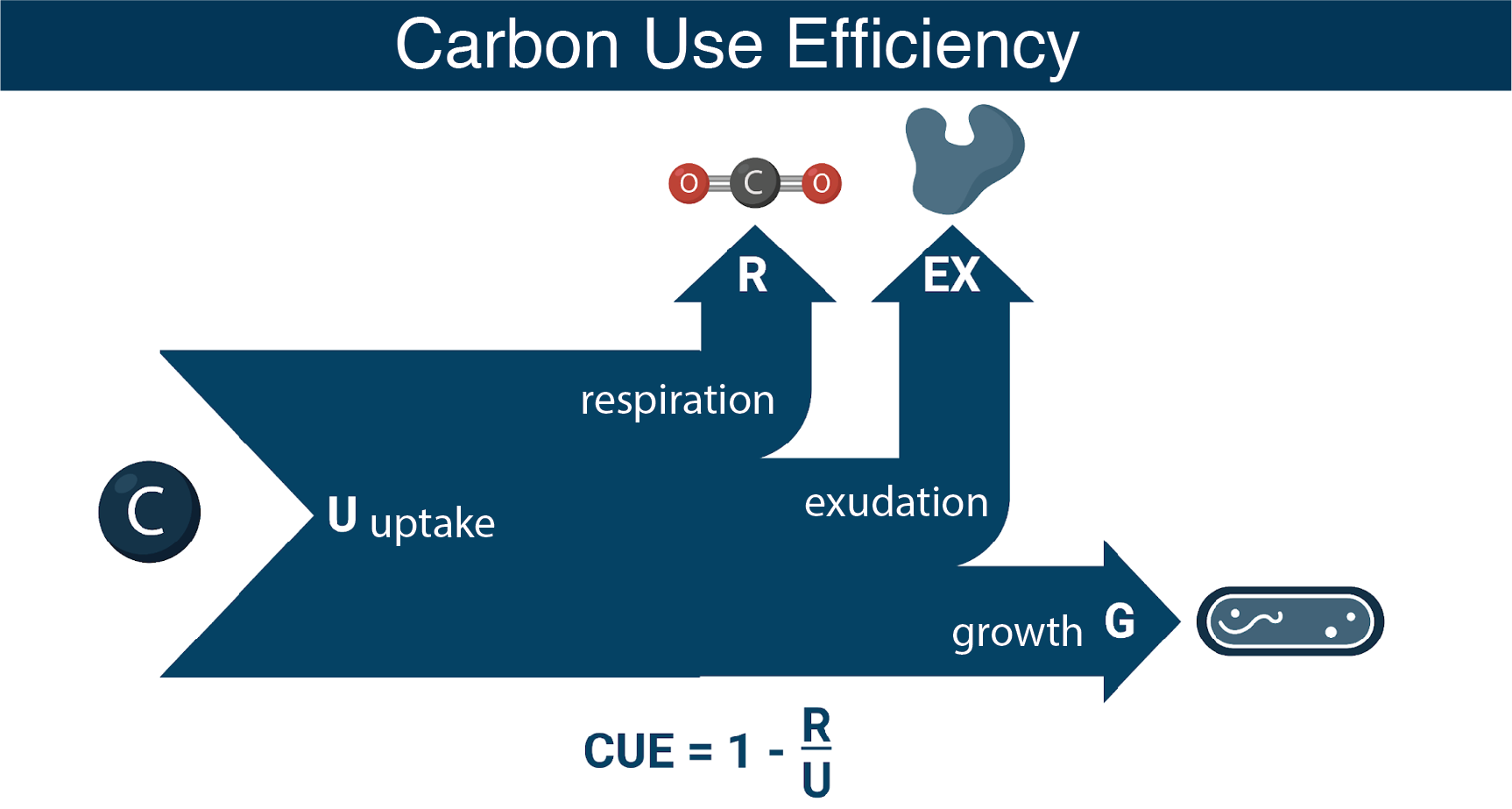
Bacterial Central Carbon Metabolism as a Predictor of CUE
The model bacteria Alteromonas macleodii MIT1002 and Ruegeria pomeroyi DSS-3 represent two distinct metabolic strategies common among marine bacteria: the preferential uptake and catabolism of sugars as growth substrates (glycolytic; substrate carbon is processed primarily via glycolysis) and the preferential uptake of organic acids (gluconeogenic; substrate carbon is processed primarily by non-glycolytic pathways) (Gralka et al., 2022). Building knowledge of how these fundamental differences in core metabolism direct carbon into biomass building versus respiratory pathways is contributing to a decades-long endeavor in marine microbial ecology to understand and predict bacterial CUE.
Experimental measurements of CUE during growth of the model strains on glycolytic versus gluconeogenic substrates, including cellular allocation of transcripts and proteins, are being integrated with flux-balance analysis (FBA) modeling of metabolic networks to understand the role of bacterial metabolic constraints and substrate preferences in determining the fate of labile carbon in the surface ocean. In order to ensure accurate predictions from FBA, the genome-scale metabolic models (GEMs) for Alteromonas macleodii MIT1002 and Ruegeria pomeroyi DSS-3 must be curated, a labor-intensive process that can be shortened by focusing exclusively on central carbon metabolism.
The Alteromonas Pangenome Metabolic Network
A pangenome consisting of the full complement of genes carried by members of the genus Alteromonas is being assembled to address gene pool filtering into species and ecotypes. A. macleodii MIT1002 is a focal species of this effort. It was isolated from a Prochlorococcus NATL2A culture (Biller et al. 2015) and acts as a “helper strain” for the cyanobacterium to reduce physiological stress (for example, from reactive oxygen species or DNA damage) (Coe et al. 2016). This project uses the anvi’o platform to construct a curated pangenome of Alteromonas (assembled from 78 isolates and 258 metagenome assembled genomes) using the newly developed Digital Microbe concept. Pangenomes (and genomes) can then be converted into chemical reaction stoichiometries using a new anvi’o tool. Network analysis with a single pangenome topology as its foundation is being used to characterize individual members of the species as a function of protein relatedness and frequency in genomes. Future research will convert the metabolic stoichiometries into working FBA models and dynamic FBA (dFBA) models at the pangenome or species level, continuing model systems tool development for better understanding marine microbes, including ecological and evolutionary implications for carbon use efficiency.
References
Gralka, M., S. Pollak, and O. X. Cordero. 2022. Fundamental metabolic strategies of heterotrophic bacteria. bioRxiv.
Biller, S. J., A. Coe, A.-B. Martin-Cuadrado, and S. W. Chisholm. 2015. Draft genome sequence of Alteromonas macleodii strain MIT1002, isolated from an enrichment culture of the marine cyanobacterium Prochlorococcus. Genome Announcements 3:e00967-15.
Coe, A., J. Ghizzoni, K. LeGault, S. Biller, S. E. Roggensack, and S. W. Chisholm. 2016. Survival of Prochlorococcus in extended darkness. Limnology and Oceanography 61:1375-1388.
